Projects I Worked On
During My Bachelor's
Insilco study on human delta sarcoglycan protein involved in Limb Girdle Muscular Dystrophy.
Read More..
- Conducted modeling of Delta Sarcoglycan protein using I-Tasser and validated the results using SAVES and Rampage. I then improved the structures using SPDBV and What-if software.
- Acquired a comprehensive understanding of the structure of proteins, including primary, secondary, and tertiary structures, and their impact on protein function.
- Investigated the interactions of proteins with other proteins, comparing wild-type and mutant forms using Haadock and PDBsum software.

Functional impact on G108R mutations in PPT1 causing infantile neuronal ceroid lipofuscinosis: A molecular dynamics Study
Read More..
- Protein structure modeling and validation, leveraging various servers to achieve accurate results.
- Studied the impact of various mutations on the PPT protein using SNP servers, gaining insights into the effects of genetic variation on protein function.
- Induced mutations in the wild-type PPT protein to study the resulting effects on stability and other key protein properties. I then performed dynamic studies using GROMACS, a widely-used molecular dynamics simulation software.
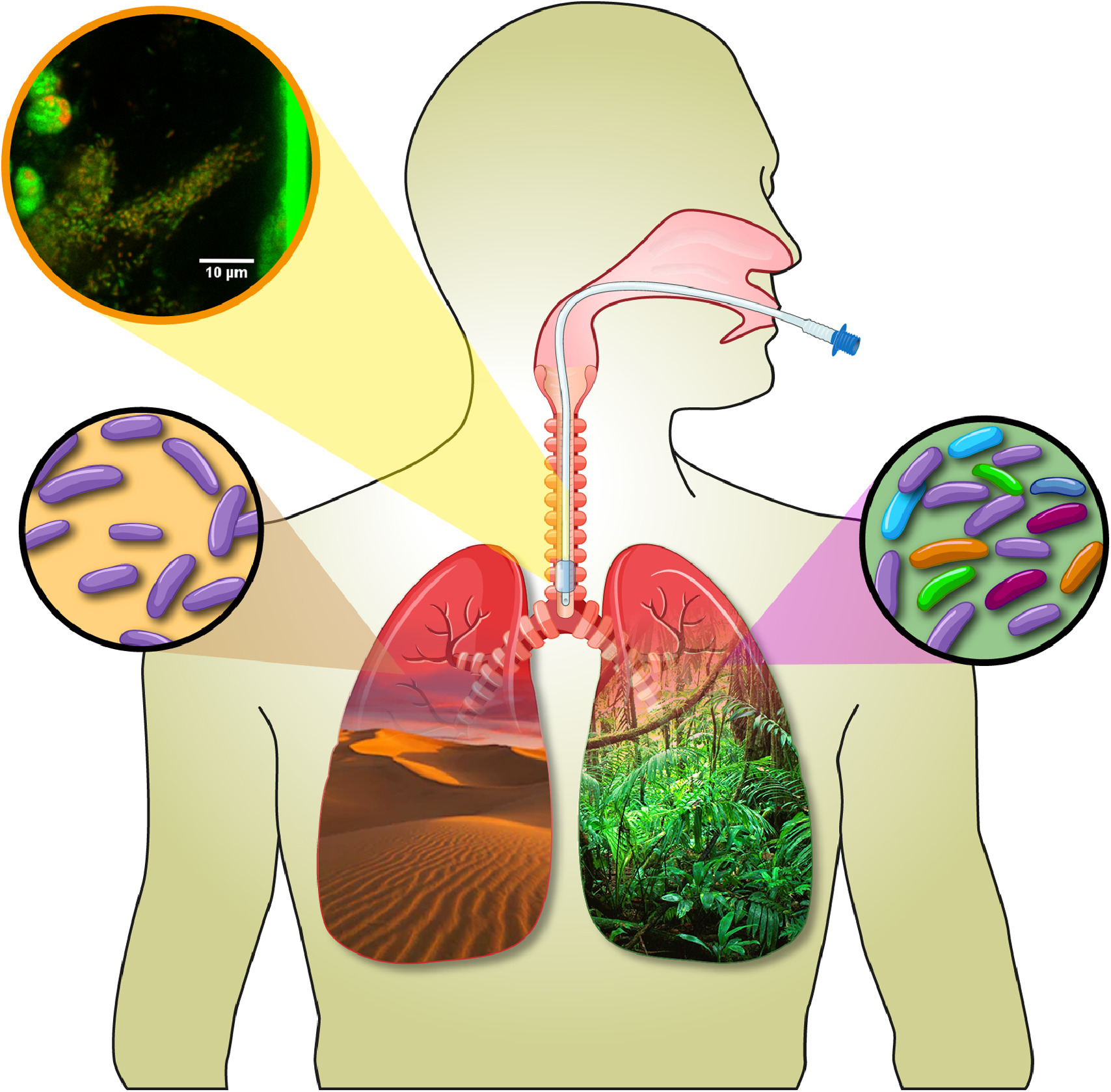
Enzymatic and antibiotic degradation of Bacterial biofilm in Ventilator Associated pneumonia.
Read More..
- Studied the formation of biofilms on endotracheal tubes of patients with Ventilator-Associated Pneumonia (VAP), deepening my understanding of the microbial ecology of this disease.
- Conducted extensive research into the development of various combinations of enzymes and antibiotics to effectively degrade these biofilms, exploring innovative approaches to treating and preventing VAP in affected patients.
During My Tenure in Boltzmann Labs
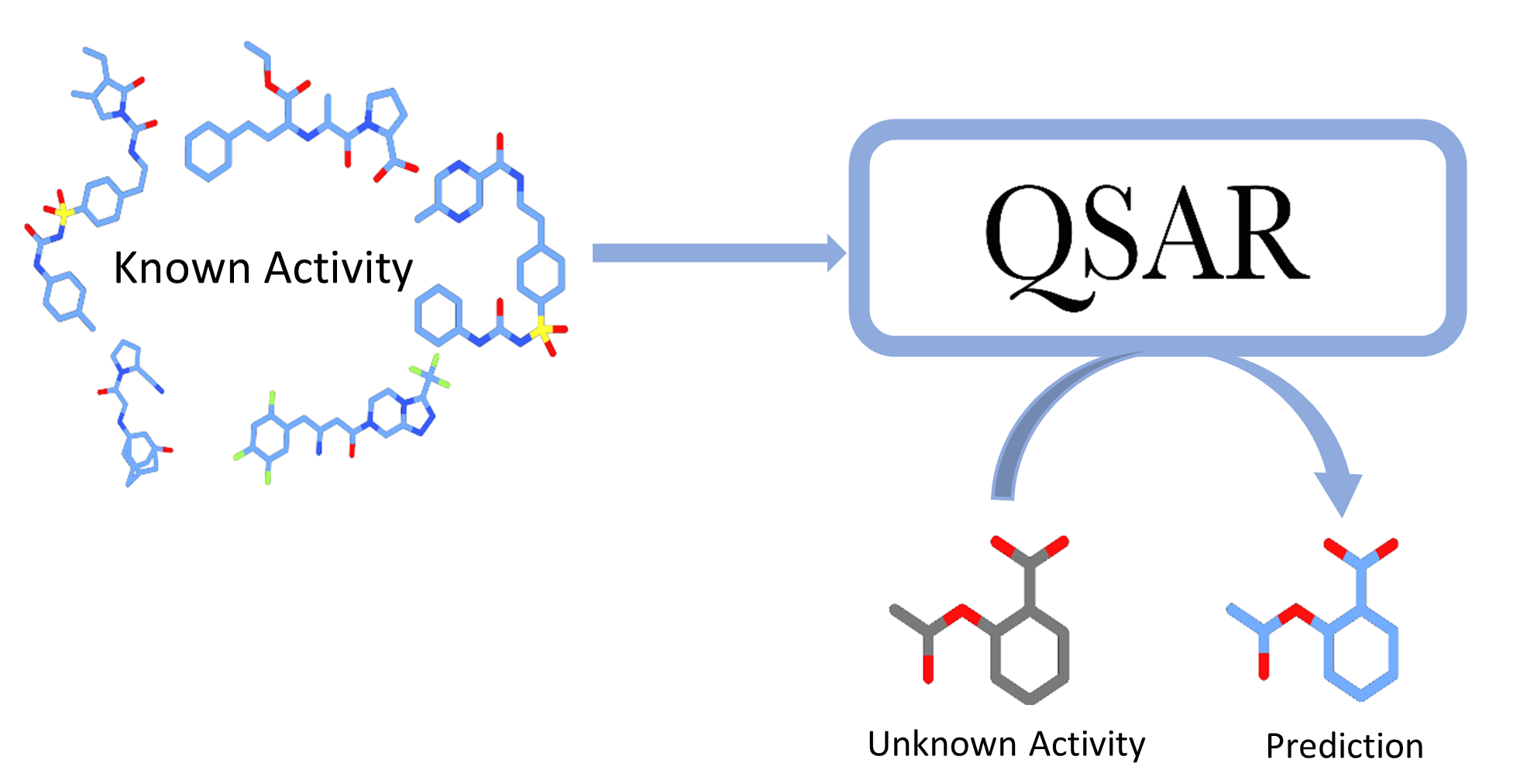
- Developed robust web scraping scripts capable of efficiently extracting data from a variety of public resources, using unique IDs as input, and curating the resulting data for use in Quantitative Structure-Activity Relationship (QSAR) modeling.
- Utilized both automated machine learning (AutoML) and graph-based deep learning models to develop highly accurate QSAR models capable of predicting complex chemical interactions. I have also incorporated interpretability and uncertainty metrics into these models, improving their overall reliability and usefulness.
- Deployed the resulting models using FastAPI, creating a user-friendly and highly accessible interface for scientists and other stakeholders in the chemical and pharmaceutical industries.
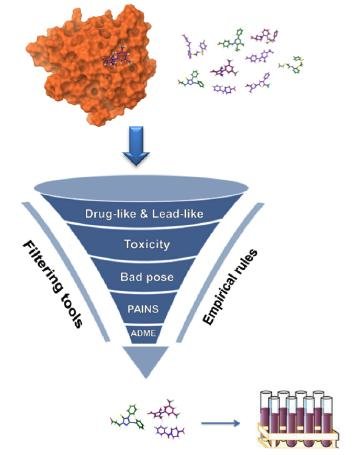
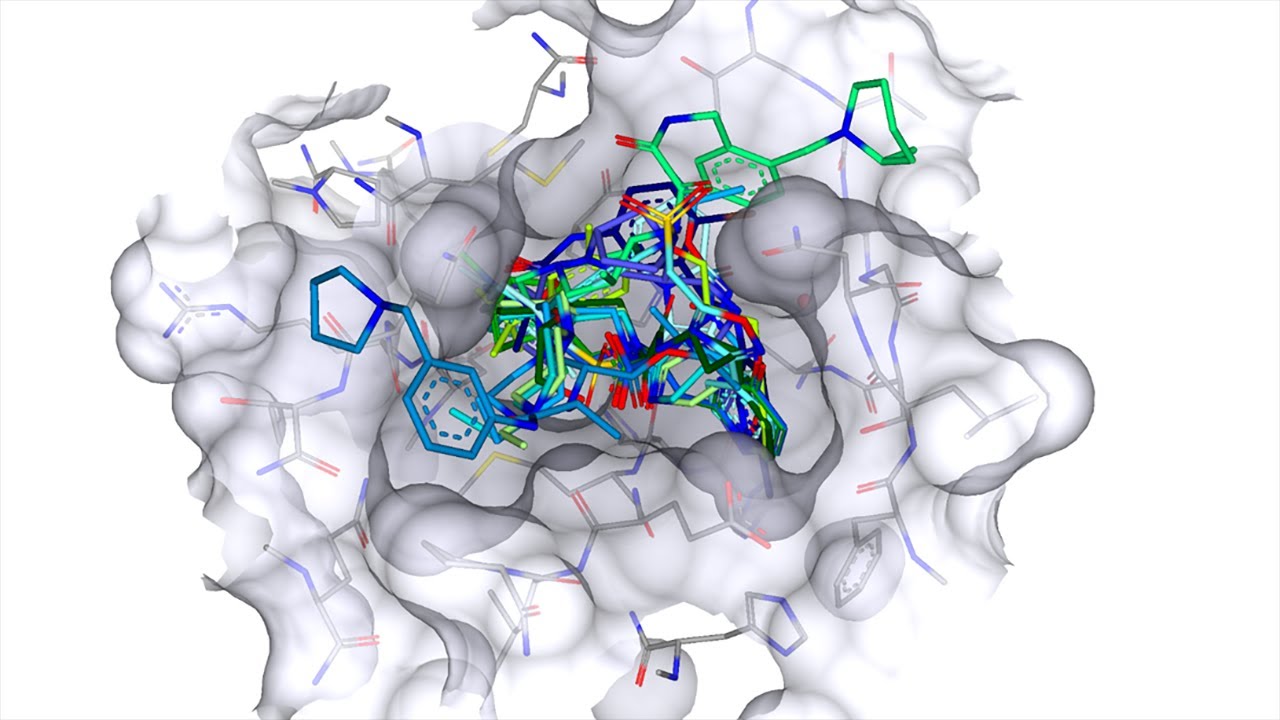
Automated virtual Screening and Validation of Small molecules: ADMET and Molecular docking
Read More..
- Developed an in-house package for calculating rule-based properties, enabling efficient and accurate analysis of complex chemical structures.
- Utilized advanced AI/ML/DL frameworks to develop predictive models for ADME and toxicity properties, providing critical insights into the safety and efficacy of potential drug compounds.
- Developed a powerful re-scoring model for predicting the affinity of drugs against proteins, using cutting-edge RESENT architectures to achieve superior accuracy and reliability.
- Created a sophisticated docking pipeline that can perform docking calculations based on user-supplied proteins and smiles. The pipeline returns detailed information about the resulting interactions, pose, efficiency, and ideal pKi values, enabling rapid analysis and optimization of potential drug candidates.
- Developed advanced strategies for exploring the chemical space in order to optimize QSAR modeling. By leveraging techniques such as active learning and uncertainty sampling, I am able to efficiently identify key areas of the chemical space that are most likely to yield high-quality data and accurate modeling results.
- Implemented a sophisticated surrogate docking pipeline that is capable of predicting the affinity of drug compounds without actually performing docking calculations. This approach enables faster and more efficient analysis of potential drug candidates, while still providing accurate and reliable insights into their properties.
Current Work
- Worked closely with DevOps teams to promote dvanced omics pipelines using Nextflow pipelines to a GCP SaaS architecture, and have been instrumental in creating automated deployment and testing environments using PyTest and Unit.
- Assisted in troubleshooting these pipelines, enabling efficient and accurate analysis of complex biological data.
- Maintained rigorous version control of pipelines on Github using a submodule strategy, and have taken steps to ensure the reliability and reproducibility of analyses by maintaining docker image versions.
- Designed a dynamic and user-friendly GUI for biologists to access and utilize all available pipelines with just a few clicks, using Flask. I have also hosted this interface on Cloud Run, and have integrated it with BQ and CloudSQL for enhanced functionality and flexibility.
- As a subject matter expert (SME), I have played a key role in designing wireframes and user flows for user interfaces, ensuring that they meet the needs of both clients and end users.
- I have also been responsible for developing and implementing data parsing strategies that leverage automation where possible, ensuring that data is processed quickly and accurately.
- In addition to my technical responsibilities, I have provided other technical support to programmers, helping them understand client requirements and guiding them as they develop software solutions.
- I have also tracked and documented progress and processes throughout the development lifecycle, ensuring that all stakeholders are informed and engaged and that projects stay on track.